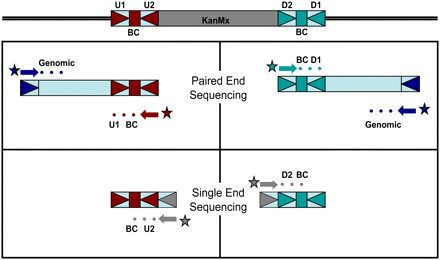
Schematic showing sequencing strategy for re-characterization of barcode and common priming sequences. (U1, U2/D1, D2) Common priming sites for uptag/downtag barcodes. (BC) Barcode. (Top panels) We used a paired-end sequencing reaction to identify both genomic position (from one read) and the barcodes and U1/D1 sequences (from the second read). (Bottom panels) In an additional sequencing reaction, we identified the barcodes and U2/D2 sequences in a single Illumina sequencing read by using a primer with homology with the KanMX4 cassette and flanking the U2/D2 sequences (shown in gray). (Colored circles) The bases that are being sequenced; (colored arrows) the primers used in the sequencing reaction; (square) the uptag barcode; (light-blue square) the downtag barcode. (Triangles flanking the colored boxes) The common primers; (dark blue triangle) the ligated adaptor sequence used to sequence the genomic DNA flanking the cassette.











