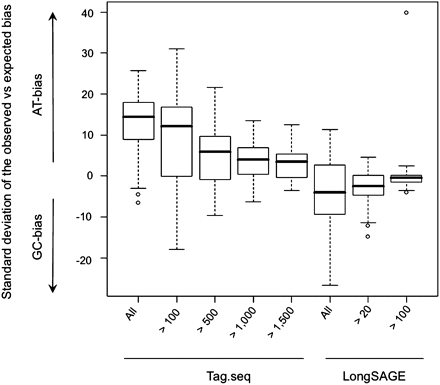
GC-bias of Tag-seq and LongSAGE libraries was calculated in units of the number of SDs by which the observed bias differed from the expected bias (see text). Positive units represent libraries with more AT-rich tag sequences than expected (AT-bias), while negative units represent libraries with more GC-rich tag sequences than expected (GC-bias). Calculated bias is shown for all quality filtered Tag-seq and all LongSAGE tag sequences, at increasing thresholds of tag expression (x-axis).











