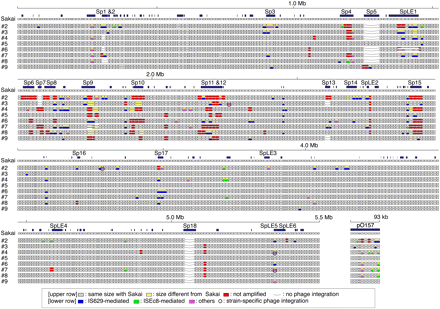
Systematic SSSP analysis results for eight O157 strains. (Upper row) For each strain, WGPS data (Ohnishi et al. 2002) are presented; (lower row) the results of the SSSP analysis. In the WGPS data, segments that yielded PCR products with lengths identical to or different from O157 Sakai are represented by gray or yellow rectangles, respectively; (red) those that were not amplified. In this study, we analyzed all of the yellow segments, which contain SSSPs. (Blue) Segments found to contain IS629-related genomic rearrangements; (light green) those containing ISEc8-related rearrangements; (magenta) segments containing other types of structural changes; of these, segments containing strain-specific phage integrations are circled; (blue rectangles) genomic regions >500 bp in length that are not present in K-12 (Sakai-specific regions or S-loops). Loci lacking prophages (Sp1–Sp18) or prophage-like integrative elements (SpLE1–SpLE6) are indicated by a line.











