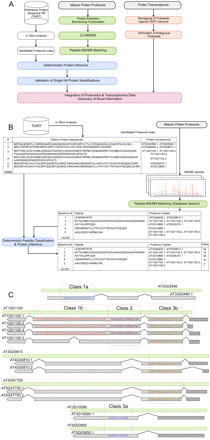
Overall workflow and peptide classification scheme. (A) Our workflow integrates the in silico analysis of the Arabidopsis reference protein database (TAIR7; Supplemental Fig. S3) to generate an identifiable proteome index (open boxes); the extraction, biochemical processing, and digestion of pollen proteins followed by mass spectrometric analysis and identification of peptides (green boxes); the manual validation of single hit proteins following deterministic peptide classification and protein inference (blue boxes); the reanalysis of transcriptomics data after remapping of the probe sets versus the TAIR7 genome and elimination of ambiguous probe sets (orange boxes); and finally the integration of proteomics and transcriptomics data allowing for discovery of novel information (pink box). (B) In silico analysis of TAIR7 allows definition of the identifiable proteome and the protein sequence–protein accession–gene model relationships. Comparison of the database search results with this identifiable proteome index (29,988 distinct protein sequences) allows us to classify each experimentally observed peptide (143,187) according to its information content and to subsequently report a conservative list of unambiguous protein identifications, as well as a likely list of proteins identified by ambiguous peptides. (C) Schematic visualization of our classification into five evidence classes. We show examples of experimentally observed peptides of class 1a (e.g., QNASYQAGQATGQTK, which unambiguously identifies AT5G65880.1); class 1b (e.g., NVTDLIMNVGAGGGGGAPVAAAAPAAGGGAAAAPAAEEK, which could imply three proteins with identical sequence that may only differ in their 5′ or 3′ UTRs [only the 5′ UTRs are represented as dark gray boxes], namely AT1G01100.1, AT1G01100.2, and AT1G01100.4 of the gene model AT1G01100, but not the splice variant AT1G0110.3, which has a different protein sequence); class 2 (e.g., AAGVSIESYWPMLFAK, which implies all splice variants of gene model AT1G01100 [in this case two distinct protein sequences]; class 3a (e.g., EGDILTLLESER, which unambiguously identifies one protein sequence that can be encoded by the distinct gene models AT3G10090.1 and AT5G03850.1). Finally, class 3b gathers peptides pointing to different protein sequences encoded by different gene models (ambiguous protein identifications).











