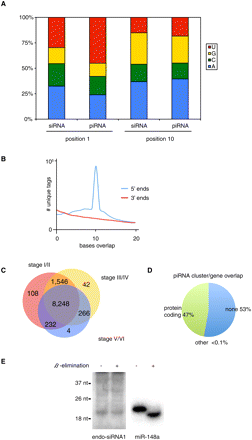
(A) Base composition at positions 1 and 10 was analyzed separately for piRNAs and 20–24 nt RNAs (siRNAs). (B) Analysis of 5′ and 3′ base overlap for unique tags. (C) Overlap of piRNA populations cloned from different stages of oogenesis. We considered only unique tags with ≥10 reads. (D) Overlap between piRNA clusters and annotated genes. (E) Identification of endo-siRNA in Xenopus tropicalis. The expression of endo-siRNA1 (5′-ACGGCCGGGGGCATTCGTATT-3′) was validated using Northern blotting after β-elimination, to assay for 2′O-methyl-modified 3′ nucleotides. miR-148a was used as a control for β-elimination as well as a loading control.











