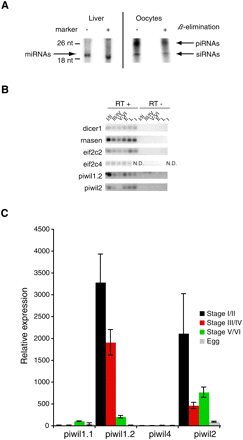
Expression of small RNAs and core protein components in X. tropicalis. (A) Total RNA was isolated from X. tropicalis adult liver and stage I and stage II oocytes. RNA was size-selected using the miRVana kit. Ten micrograms of this RNA was subjected to β-elimination or not as indicated and analyzed on a denaturing gel after 5′ end-labeling. Arrows indicate RNA that likely represent miRNAs, siRNAs, and piRNAs. (B,C) The expression of core components of small RNA pathways in Xenopus oocytes, eggs, and somatic tissues was assayed using RT-PCR and qRT-PCR. For each experiment equivalent oocyte total RNA was reverse transcribed. E, egg; L, liver; I, intestine; N.D., not done. (B) RT-PCR experiment. Control experiment without the addition of reverse transcriptase. RT-PCR primers are listed in Supplemental material, Armisen_SupData4.xls. (C) qRT-PCR for the four Piwi-related genes described in X. tropicalis.











