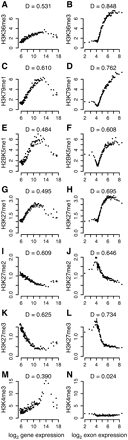
Histone modifications are highly dependent on exon-expression level. Histone modification signals over gene bodies (A,C,E,G,I,K,M) and internal exons (B,D,F,H,J,L,N) related to gene- and exon-expression bins, respectively, for H3K36me3 (A,B), H3K79me1 (C,D), H2BK5me1 (E,F), H3K27me1 (G,H), H3K27me2 (I,J), H3K27me3 (K,L), and H3K4me3 (M,N). The Hoeffding's D statistic (indicated above each plot) measures the dependency of histone modification signal on expression level (see Methods for details).











