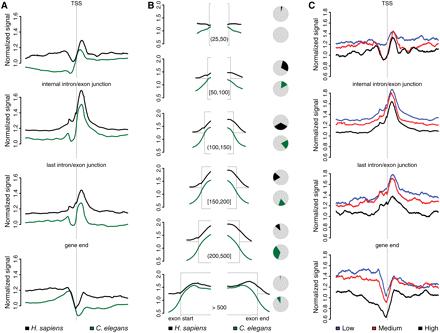
Nucleosomes are well positioned at internal exons. (A) Footprints of normalized nucleosome signal in human T-cells and C. elegans in a ±1-kb window. Signals were normalized for the total number of sequenced bases and genome size. The windows are centered, from top to bottom, on transcription start sites, intron/exon junctions of internal exons, intron/exon junctions of last exons, and 3′ ends of genes. (B) Partial footprints (left) of nucleosome signal in human (black) and C. elegans (green) at internal exon starts and ends split into six groups according to exon length (bp intervals given in brackets). In the pie charts (right), the percentage of the total number of exons in each exon size category is shown for human (black) and C. elegans (green). Included exons have flanking introns that are at least 100-bp long. (C) Footprints of human nucleosome signals for the same exon categories as in A but divided according to gene expression.











