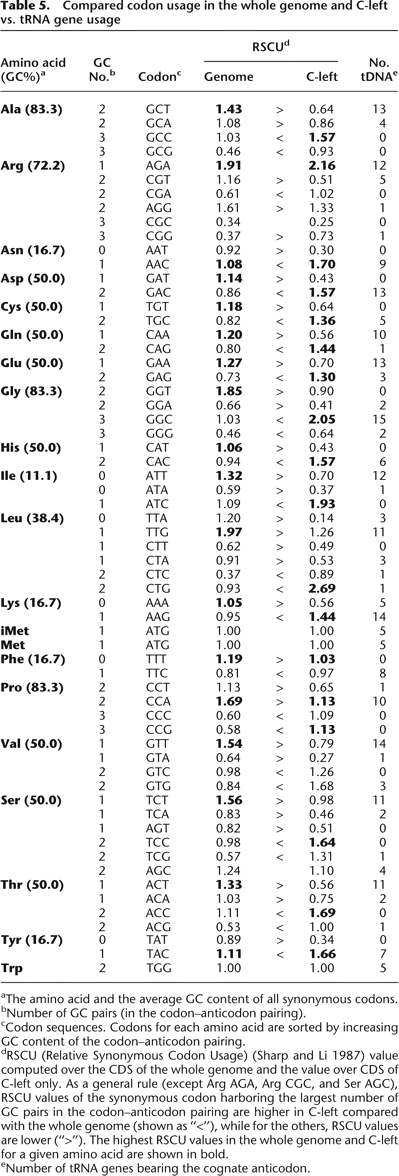
Compared codon usage in the whole genome and C-left vs. tRNA gene usage
Click on table to view larger version.
-
aThe amino acid and the average GC content of all synonymous codons.
-
bNumber of GC pairs (in the codon–anticodon pairing).
-
cCodon sequences. Codons for each amino acid are sorted by increasing GC content of the codon–anticodon pairing.
-
dRSCU (Relative Synonymous Codon Usage) (Sharp and Li 1987) value computed over the CDS of the whole genome and the value over CDS of C-left only. As a general rule (except Arg AGA, Arg CGC, and Ser AGC), RSCU values of the synonymous codon harboring the largest number of GC pairs in the codon–anticodon pairing are higher in C-left compared with the whole genome (shown as “<”), while for the others, RSCU values are lower (“>”). The highest RSCU values in the whole genome and C-left for a given amino acid are shown in bold.
-
eNumber of tRNA genes bearing the cognate anticodon.











