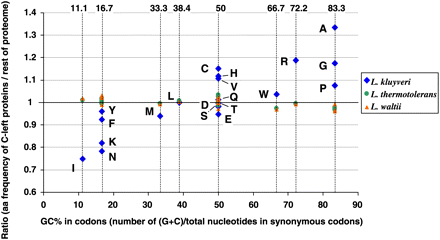
Biased amino acid composition of proteins encoded by C-left. The ratio between the amino acid frequency of proteins encoded by C-left of L. kluyveri and of those encoded by the rest of the genome is plotted along the average nucleotide composition of all synonymous codons for the corresponding amino acid (exact values are on the top of the graph). Amino acids are identified by single-letter code next to blue diamonds. Residues located >1.0 are overrepresented in C-left compared with the rest of the genome; residues <1.0 are underrepresented. Extreme cases are I (isoleucine), which is used 25% less (ratio = 0.75) in C-left than in the rest of the genome, and A (alanine), which is used 33% more (ratio = 1.33). The same analysis was performed for the proteins of L. thermotolerans (green dots) and L. waltii (orange triangles), which are considered orthologous to the C-left proteins of L. kluyveri.











