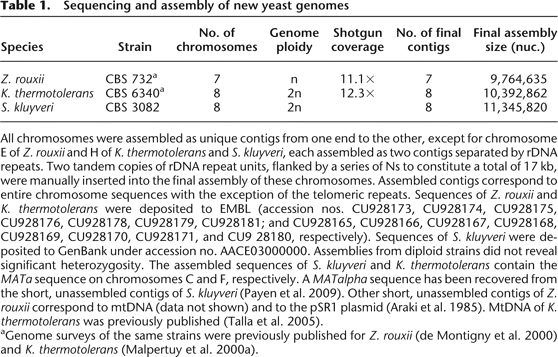
Sequencing and assembly of new yeast genomes
Click on table to view larger version.
-
All chromosomes were assembled as unique contigs from one end to the other, except for chromosome E of Z. rouxii and H of K. thermotolerans and S. kluyveri, each assembled as two contigs separated by rDNA repeats. Two tandem copies of rDNA repeat units, flanked by a series of Ns to constitute a total of 17 kb, were manually inserted into the final assembly of these chromosomes. Assembled contigs correspond to entire chromosome sequences with the exception of the telomeric repeats. Sequences of Z. rouxii and K. thermotolerans were deposited to EMBL (accession nos. CU928173, CU928174, CU928175, CU928176, CU928178, CU928179, CU928181; and CU928165, CU928166, CU928167, CU928168, CU928169, CU928170, CU928171, and CU9 28180, respectively). Sequences of S. kluyveri were deposited to GenBank under accession no. AACE03000000. Assemblies from diploid strains did not reveal significant heterozygosity. The assembled sequences of S. kluyveri and K. thermotolerans contain the MATa sequence on chromosomes C and F, respectively. A MATalpha sequence has been recovered from the short, unassembled contigs of S. kluyveri (Payen et al. 2009). Other short, unassembled contigs of Z. rouxii correspond to mtDNA (data not shown) and to the pSR1 plasmid (Araki et al. 1985). MtDNA of K. thermotolerans was previously published (Talla et al. 2005).
-
aGenome surveys of the same strains were previously published for Z. rouxii (de Montigny et al. 2000) and K. thermotolerans (Malpertuy et al. 2000a).











