Bioinformatic analysis of 47 P. aeruginosa LESB58 genes essential for lung infection as identified by PCR-based screening of 9216 STM mutants after passage through the chronic rat lung agar bead infection model
Click on table to view larger version.
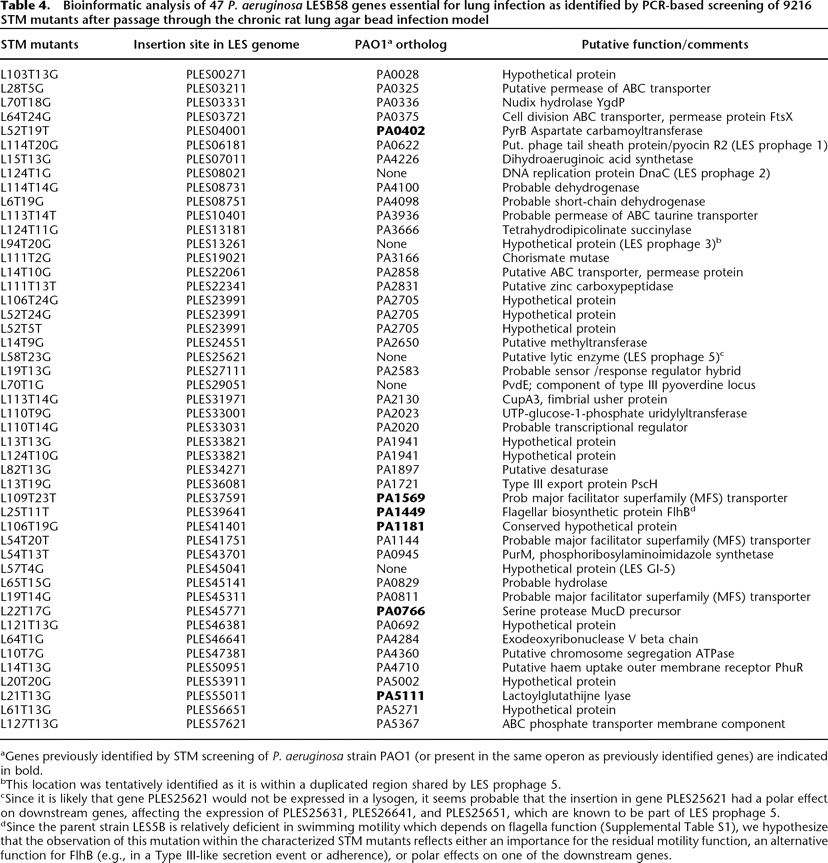
aGenes previously identified by STM screening of P. aeruginosa strain PAO1 (or present in the same operon as previously identified genes) are indicated in bold.
bThis location was tentatively identified as it is within a duplicated region shared by LES prophage 5.
cSince it is likely that gene PLES25621 would not be expressed in a lysogen, it seems probable that the insertion in gene PLES25621 had a polar effect on downstream genes, affecting the expression of PLES25631, PLES26641, and PLES25651, which are known to be part of LES prophage 5.
dSince the parent strain LES5B is relatively deficient in swimming motility which depends on flagella function (Supplemental Table S1), we hypothesize that the observation of this mutation within the characterized STM mutants reflects either an importance for the residual motility function, an alternative function for FlhB (e.g., in a Type III-like secretion event or adherence), or polar effects on one of the downstream genes.











