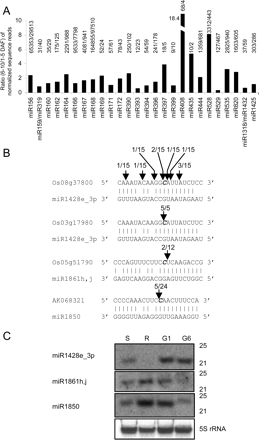
(A) Ratio (6–10 DAF/1-5 DAF) of the normalized miRNA sequence reads of the known miRNA families with at least 10 reads in total in the Illumina data set. The raw sequence reads of each miRNA family is shown on top of the bars with numerator and denominator representing number of reads in 6–10 DAF and 1–5 DAF grains, respectively. (B) Mapping of miRNA-guided cleavage sites in predicted target genes. Predicted cleavage sites are indicated by a bold italic nucleotide at position 10 relative to the 5′ end of the miRNA. Mapped cleavage positions are indicated by arrows with the frequency amongst 5′ RACE clones sequenced. (C) Northern blot detection of expression of miRNAs in different tissues. Uniform RNA loading was demonstrated by reprobing blots with 5S rRNA (representative blot shown). (S) Shoots; (R) roots; (G1) 1–5 DAF grains; (G6) 6–10 DAF grains.











