Influence of duplication on substitution parameters
Click on table to view larger version.
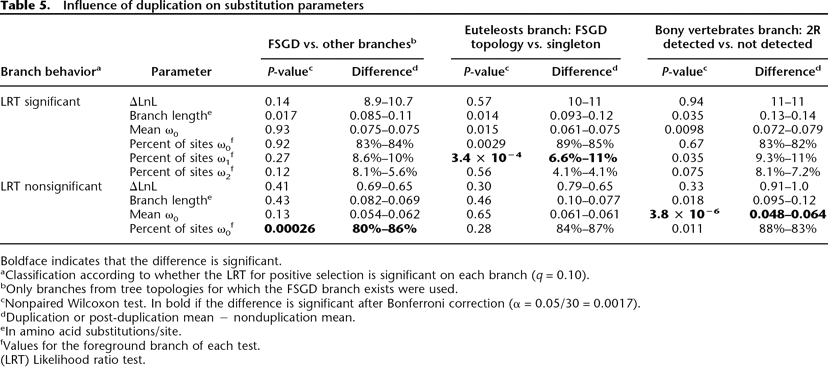
Boldface indicates that the difference is significant.
aClassification according to whether the LRT for positive selection is significant on each branch (q = 0.10).
bOnly branches from tree topologies for which the FSGD branch exists were used.
cNonpaired Wilcoxon test. In bold if the difference is significant after Bonferroni correction (α = 0.05/30 = 0.0017).
dDuplication or post-duplication mean − nonduplication mean.
eIn amino acid substitutions/site.
fValues for the foreground branch of each test.
(LRT) Likelihood ratio test.











