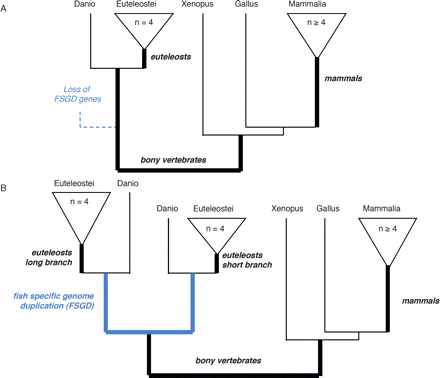
Tree topologies studied. Schematic representation of the tree topologies selected. (Black) speciation branches; (blue) duplication branches. Branches in bold were used as “foreground branches” in branch-site tests for positive selection. Bold italics indicate branch names used in the text and the tables. (A) “Singleton” tree type, with no duplication allowed. The dotted line represents gene loss after whole genome duplication. (B) “Fish-specific whole genome duplication” tree type. Duplication on other branches may be excluded or allowed. Because differences in dN/dS ratio have been reported between paralogs (Brunet et al. 2006; Scannell and Wolfe 2007), with higher dN/dS in the fast evolving paralog, we distinguish the longer and shorter branch after duplication, based on the PhyML protein phylogeny.











