A chromosome comparison of segmental duplications—DupMasker vs. WGAC
Click on table to view larger version.
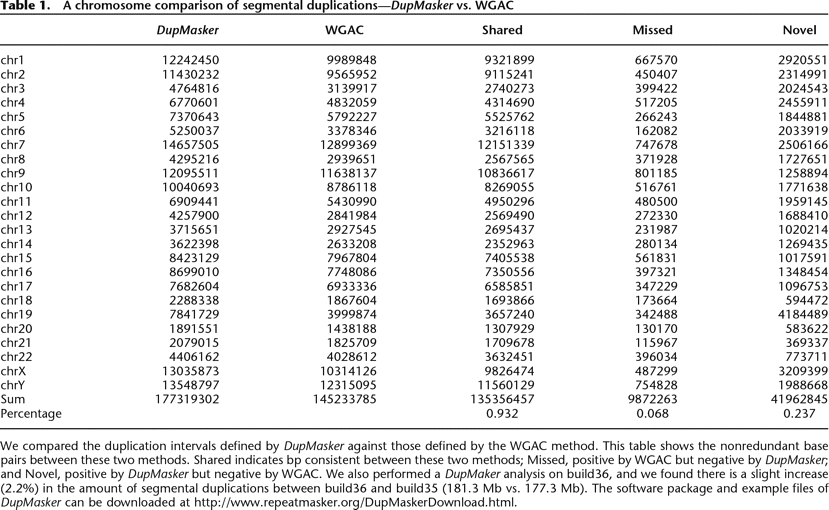
We compared the duplication intervals defined by DupMasker against those defined by the WGAC method. This table shows the nonredundant base pairs between these two methods. Shared indicates bp consistent between these two methods; Missed, positive by WGAC but negative by DupMasker; and Novel, positive by DupMasker but negative by WGAC. We also performed a DupMaker analysis on build36, and we found there is a slight increase (2.2%) in the amount of segmental duplications between build36 and build35 (181.3 Mb vs. 177.3 Mb). The software package and example files of DupMasker can be downloaded at http://www.repeatmasker.org/DupMaskerDownload.html.











