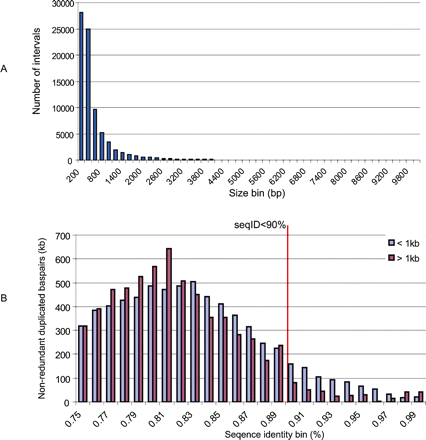
The size and sequence identity distribution of “novel” duplications. (A) The length distribution of DupMasker duplications not detected by WGAC (termed “novel” SDs) reveals that the majority (99% by number of intervals, 91% by base pair) of these intervals are small fragments (size <1 kb). (B) We found 52.3% (21.9 Mb) of these small intervals are common repeats due to imprecision of boundary definition within repeat-rich regions. We performed a modified WGAC analysis using a relaxed threshold (require nonrepeat alignment ≥100 bp and sequence identity ≥75%) on these “novel” SDs. The analysis revealed alignments for 31% (13.1/41.96 Mb) of these “novel” SDs. Among the 13.1-Mb alignments, 97.7% (12.8/13.1 Mb) represent either small (size <1 kbp) or relatively ancient duplications (sequence identity <90%).











