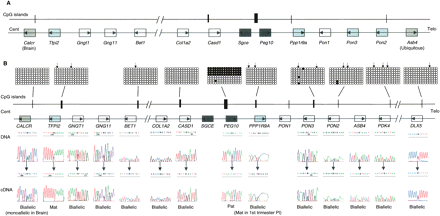
Genomic comparison between proximal mouse chromosome 6 and human chromosome 7q21 imprinted domains. (A) Schematic representation of mouse Peg10 domain, showing the relative organization of genes and CpG islands. (B) Schematic representation of the orthologous human domain. The methylation status of all human promoter CpG islands was examined in liver-, muscle-, lymphocyte-, and placenta-derived DNA. Methylation patterns were assessed first by restriction digestion of bisulfite PCR products. The positions of the restriction sites used are shown (↓). Patterns obtained by sequencing bisulfite PCR products are examples obtained from placenta-derived DNA. Similar patterns were obtained in all tissues analyzed. Each circle represents a single CpG dinucleotide on the strand, a methylated cytosine (●), or an unmethylated cytosine (◯). Only the first 15 CpG dinucleotides from each CpG island are shown. The only evidence for differential methylation was at the known DMR located between the promoters for PEG10 and SGCE; all other promoter CpG islands were hypomethylated. The imprinted expression for all genes in the cluster was analyzed in first trimester fetal tissues and term placenta. DNA sequence traces for heterozygous term placenta samples are shown for all genes, and the resulting RT-PCR sequences. Open boxes depict genes that are biallelic in all tissues throughout gestation, whereas dark gray boxes represent the ubiquitously imprinted, paternally expressed genes. Light gray boxes represent genes that are ubiquitously maternally expressed, while hatched boxes are the placental-specific maternally expressed genes. Arrows show the direction of transcription for each gene.











