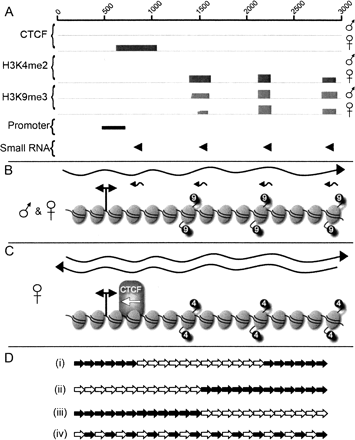
Summary of the chromatin organization and transcription of DXZ4 on the active and inactive X chromosomes and a schematic model of how this organization might be arranged at Xq23. (A) Combined summary of ChIP-chip, luciferase, and small RNA Northern data for a single 3-kb monomer. (B) Summary illustration of DXZ4 chromatin and transcripts in males and at the Xa. The wavy right-pointing black arrow represents the >3-kb sense strand transcript. Small left-pointing arrows represent small antisense RNAs. The elliptical shapes represent nucleosomes. A total of 15 nucleosomes are arbitrarily assigned to a single DXZ4 monomer based on ∼200-bp per nucleosome. The nucleosomes corresponding to the peaks of H3K9me3 signal are indicated by the “9”-labeled black circles. The bidirectional promoter is indicated by the double-headed arrow. (C) Summary illustration of DXZ4 chromatin and transcripts as determined for the Xi. The left- and right-facing wavy arrows represent the sense and antisense DXZ4 transcripts. The nucleosomes corresponding to the peaks of H3K4me2 signal are indicated by the “4”-labeled black circles. The position of CTCF binding is indicated by the CTCF labeled shape, and the left-pointing white arrow represents the direction of promoter–enhancer interference activity. (D) Schematic representation of possible Xi arrangement of H3K4me2 defined monomers (white arrows) and H3K9me3 monomers (black arrows). Each arrow represents a full 3-kb monomer. Only 24 of a potential 50–100 monomers were used for illustrative purposes.











