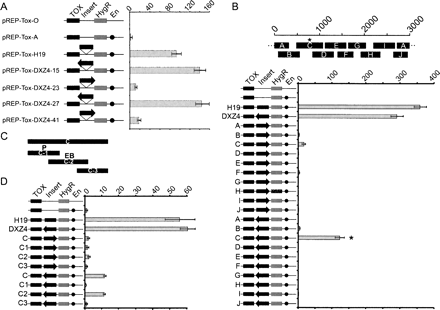
DXZ4 unidirectionally interferes with promoter–enhancer communication in vitro, an attribute confined to DNA encompassing the CTCF binding site. (A) Schematic representation of the different constructs are labeled and shown on the left. The location of the diphtheria toxin-A gene (TOX), hygromycin-B resistance gene (HygR), insert (Insert), and SV40 enhancer (En) are indicated for the rectangles and circles. Insert identity is indicated to the left of each construct. The orientation of DXZ4 inserts are represented by the black arrows. The graph to the right indicates the number of colonies obtained for each construct after 2–3 wk of selection for hygromycin-B. Data represents the mean and standard error for four independent experiments. (B) A single 3-kb DXZ4 monomer is represented by the 1–3000 bp scale. The approximate location of subfragments A–J are shown below the monomer. Data obtained as for A. The asterisk highlights the subfragment that retained most of the insulator activity. (C) Representation of the subfragments of C (C1–C3) that were cloned in either orientation into pREP-ToxA. The P indicates the fragment containing the highest promoter activity, whereas the EB indicates the fragment containing enhancer-blocking activity determined below. (D) Data obtained for the C subfragments determined as for A.











