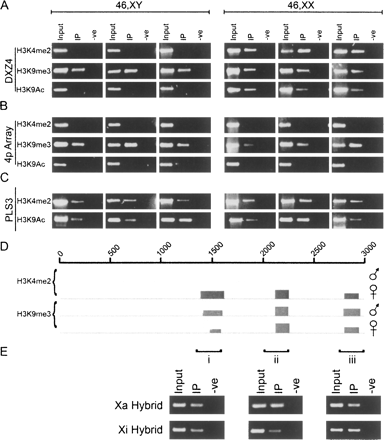
DXZ4 on the inactive X chromosome adopts a Xi-specific euchromatin-like conformation. (A) Ethidium bromide (EtBr) agarose gels of H3K4me2, H3K9me3, and H3K9Ac ChIP assessed by PCR for co-immunoprecipitation of DNA from the Xq23 macrosatellite DXZ4, and (B) an unrelated macrosatellite on chromosome 4p (4p array). Results are shown for three independent primary male and female cell lines. (C) PCR analysis of the plastin 3 promoter (PLS3). Samples include the input, experimental (IP), and control (-ve). (D) Close-up of ChIP-chip data for a single 3-kb DXZ4 monomer (0–3000 bp) with H3K4me2 and H3K9me3 in a male and female sample. The coordinates at the top of the image represent nucleotides 114,785,064–114,788,023 of Human Genome Build HG17. Immediately below the line is the NimbleGen Signal Map 1.8 output for the H3K4me2 and H3K9me3 ChIP-chip. The width of the signal indicates the extent of the hybridization. (E) H3K9me3 ChIP from the Xa and Xi hybrid assessed by PCR. The three regions analyzed by PCR are indicated by i–iii and correspond to the sites of the H3K9me3 peaks defined by ChIP-chip.











