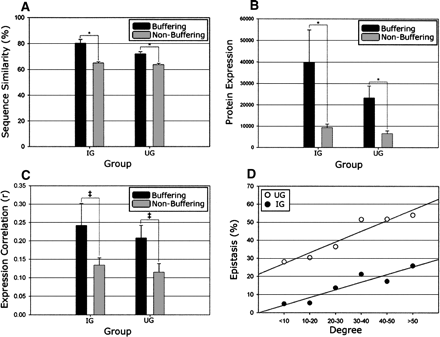
Expression and co-conservation of buffering paralogs. All relationships depict comparisons between epistatic paralogs (IG and UG) and respective non-epistatic following the removal of ribosomal proteins. (A) Buffering paralogs have significantly more co-conservation of sequence than non-buffering (P < 0.001 for IG and UG, indicated by asterisks). (B) Buffering paralogs are significantly more highly expressed than non-buffering (protein abundance depicted, same results noted for transcript abundance) and additionally exhibit a trend toward (P < 0.1, indicated by ‡) being more highly correlated throughout the cell cycle (displayed in C). (D) All WGD paralog pairs were binned based on their number of combined protein interactions (degree) within the BioGRID data set, with the respective percentage epistatic (both IG and UG) indicated, demonstrating a clear relationship between the number of physical interactions and the likelihood for epistasis under normal growth conditions.











