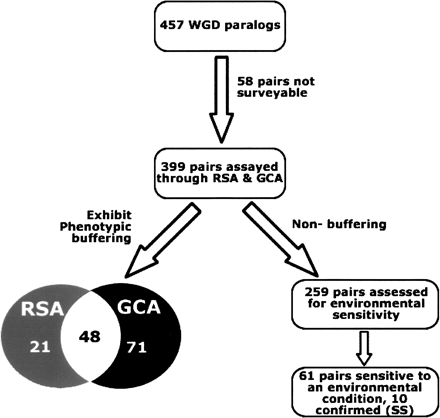
Experimental schema and summary of epistasis among WGD paralog pairs in yeast. The 399 WGD paralog pairs with viable constituent single-deletion strains (Giaever et al. 2002) were analyzed for genetic interaction using random-spore analysis (RSA) and growth-curve analysis (GCA). The overlap in terms of duplicates exhibiting significant phenotypic buffering is shown in the Venn diagram. The 259 WGD paralog pairs not exhibiting epistasis (and hence deemed to not buffer phenotypically) in rich growth media were further analyzed through environmental screening in five alternate media conditions designed to either mimic common cellular stress states or introduce competition (Giaever et al. 2002).











