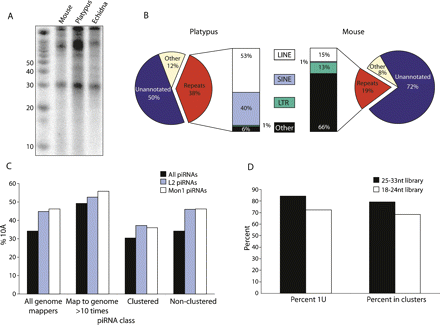
piRNAs in platypus. (A) Total RNA labeling from mouse, platypus, and echidna testis reveals a distinct ∼30-nt piRNA species in monotremes. RNA was dephosphorylated, end labeled with 32P, and run on a denaturing 15% polyacrylamide gel. Sizes of markers (left) are in nucleotides. (B) Comparison of platypus and mouse piRNA libraries. A platypus piRNA library was generated by cloning small RNAs in a 25- to 33-nt size window from total platypus testis RNA. Sequences from this library and from an equivalent mouse piRNA library (Girard et al. 2006) were classed as repeat (red), unannotated (blue), or other (beige). The repeat piRNAs were broken down into classes of repeat (bars). (C) 10A profiles for platypus piRNAs. An A at piRNA position 10 is a signature of an active piRNA amplification system. “Clustered” piRNAs are defined as those that map to the genome once and fall into one of 50 piRNA clusters (Supplemental Table 4), while “non-clustered” piRNAs are those that map to the genome once but do not fall into a cluster. 10A percentages for all piRNAs are in black, those that are derived from LINE2 elements in blue, and those from Mon1 SINE elements in white. (D) Characteristics of “small” piRNAs. “Percent 1U” includes all small RNAs that match the genome; “percent in clusters” includes only small RNAs that map only once to the genome.











