List of chicken mirtrons
Click on table to view larger version.
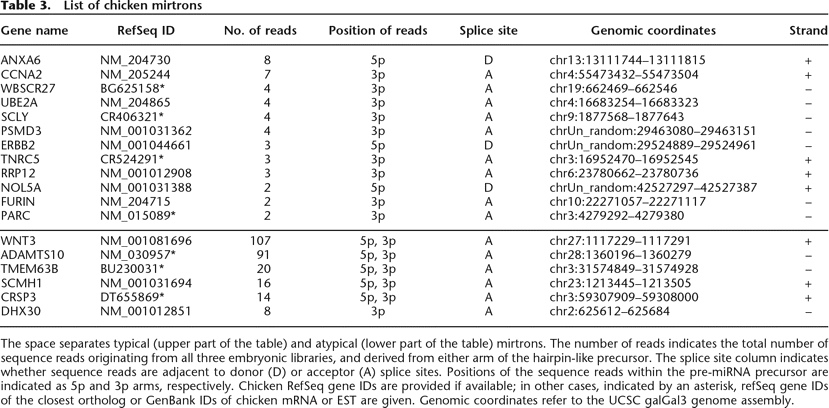
The space separates typical (upper part of the table) and atypical (lower part of the table) mirtrons. The number of reads indicates the total number of sequence reads originating from all three embryonic libraries, and derived from either arm of the hairpin-like precursor. The splice site column indicates whether sequence reads are adjacent to donor (D) or acceptor (A) splice sites. Positions of the sequence reads within the pre-miRNA precursor are indicated as 5p and 3p arms, respectively. Chicken RefSeq gene IDs are provided if available; in other cases, indicated by an asterisk, refSeq gene IDs of the closest ortholog or GenBank IDs of chicken mRNA or EST are given. Genomic coordinates refer to the UCSC galGal3 genome assembly.











