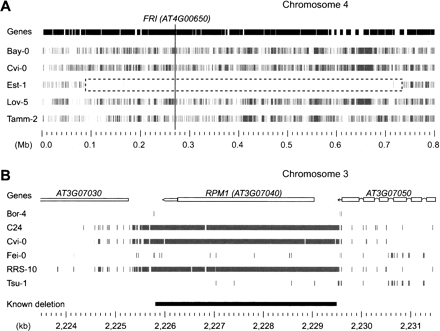
PRs reveal haplotype sharing at chromosomal and local scales. (A) Genes (top) and PRs (gray blocks beneath) for five accessions for 0.8 Mb surrounding the FRI locus. In Est-1 a region of ∼0.6 Mb (dashed black box) including FRI (vertical line) has been reported to be nearly identical to the Col-0 reference sequence but divergent in the other accessions shown (Nordborg et al. 2005; Clark et al. 2007). Only several PRs are located in the Est-1 region that is monomorphic with the tiled reference sequence. (B) Pattern of PRs for 8 kb at the RPM1 locus. The location of a 3.7-kb deletion that segregates in the A. thaliana population is as indicated at bottom (Grant et al. 1995, Shen et al. 2006). Experimental characterization revealed that the C24, Cvi-0, and RRS-10 accessions included in the current study harbored this deletion (the other accessions shown have a Col-0 like haplotype). PRs delineate the deletion as well as flanking SNPs and indels (see also Supplemental Fig. S6).











