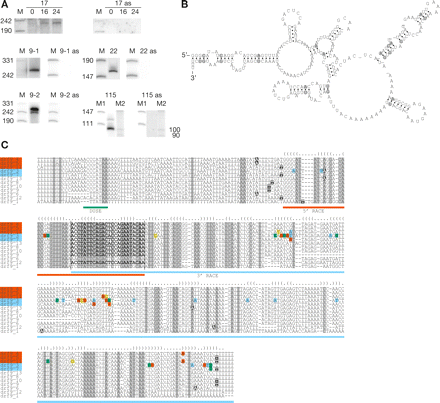
Analysis of expressed ncRNA families. (A) Northern blot analysis of families hybridized with sense and anti-sense (as) probes. drf17 was analyzed for developmental regulation on RNA extracted at 0 h (growing cells), 16 h, and 24 h. Two slightly different probes were used in the analysis of drf9 (9-1 and 9-2). (B) Consensus secondary structure, predicted by RNAalifold, of the five members matching sequences determined by RACE analysis. Compensatory base pair changes are circled. (C) Alignment of the drf9 member sequences including upstream and downstream flanking sequences. (Green box beneath sequences) Location of the DUSE sequence present in nine sequences. (Red and blue boxes beneath sequences) Sequences determined by 5′- and 3′-RACE analysis, respectively. Candidate gene names labeled with red and blue boxes indicate members matching the sequences determined by RACE analysis. (Bold letters between the vertical bars) Nucleotides complementary to the hybridization probes. (Black boxes in the alignment) Start and end point nucleotides of predictions. The predictions for drf9_3, drf9_4, and drf9_8 extend beyond the alignment for an additional 124, 226, and 111 nucleotides, respectively. (Dot-bracket notation above the alignment) The Alifold consensus structure. The notation has been slightly adapted to accommodate the full family alignment. Compensatory base pair changes in the RACE-determined sequences are indicated: (green box) a change to a GC base pair; (blue box) a change to a AT base pair; (yellow box) a change to a GT base pair; (red box) a change that disrupts base-pairing.











