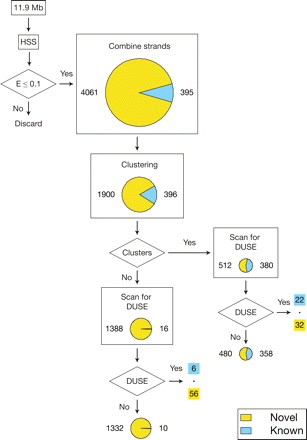
Flowchart and clustering of sequences. The six chromosomes of the D. discoideum where exons and complex repeats had been masked were searched for high-scoring segments with an expect value of at most 0.1. Overlapping high-scoring segments residing on opposite strands were combined. The predictions were filtered for presence in multi-copy families and/or the occurrence of an upstream sequence element (DUSE) within 150 nt flanking the prediction. Piecharts represent the number and character of sequences that were used for the operation. The size of the piechart is proportional to the total number of sequences. (Yellow and the associated numbers) Novel predictions; (blue and associated numbers) known or confidently predicted ncRNA genes. After combining the strands, the number of known or confidently predicted ncRNA genes increased since genes with an overlapping prediction on the template strand count as detected after the combination step.











