Table 1.
Categorization and coverage of 26 million WGS sequence traces
Click on table to view larger version.
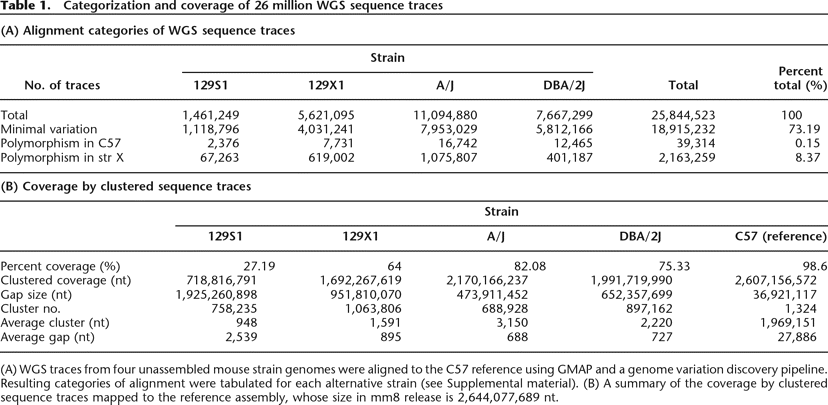
(A) WGS traces from four unassembled mouse strain genomes were aligned to the C57 reference using GMAP and a genome variation discovery pipeline. Resulting categories of alignment were tabulated for each alternative strain (see Supplemental material). (B) A summary of the coverage by clustered sequence traces mapped to the reference assembly, whose size in mm8 release is 2,644,077,689 nt.











