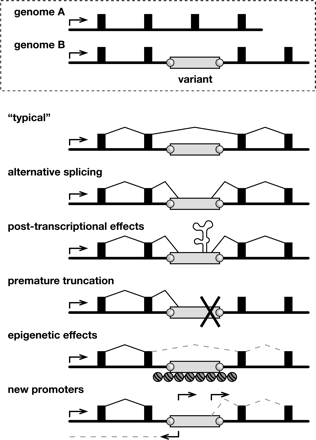
Genomic and transcriptional variation due to endogenous transposition. (Top) Schematic of allelic variants A and B at a genomic locus including a promoter (arrow), exons (filled boxes), introns (underlying black line), and a polymorphic transposon integrant (open rectangle, genome B) with target-site duplications (gray circles). (Bottom) Possible forms of transcriptional variation due to a transposon integrant. (“Typical” transcript) Because transposons are ubiquitous, a typical transcript might lack an intronic integrant by splicing between exons. (Alternative splicing) Transcripts might include portions of transposon integrants due to their internal splice donor and splice acceptor sites. (Post-transcriptional effects) Transposon sequences may introduce autoregulatory elements affecting RNA stability, intracellular compartmentalization, etc. (Premature truncation) Similar to alternative splicing, except that transcripts end prematurely due to a transcription terminator in the transposon. (Epigenetic effects) Read-through transcription may be repressed by heterochromatin, DNA methylation, and/or other epigenetic controls at transposon integrants. (New promoters) Gene expression and structure may be altered by introduction of new sense and/or antisense promoters in transposon integrants. This is the main form of transcriptional variation described in this report.











