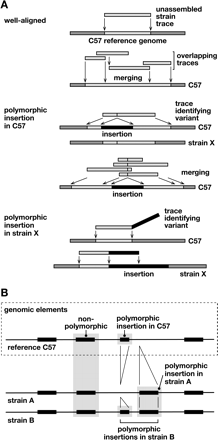
Discovery of structural variation between inbred mouse strains by WGS trace alignment. (A) Presented here is a schematic illustrating several types of alignments of WGS traces against the reference genome (Supplemental Fig. 1). (Top) “Well-aligned” traces aligned almost completely at unique locations in the C57 reference genome (light gray). Overlapping sequences were merged into a contig. (Middle) “Polymorphic insertion in C57” traces identify an indel present in the C57 reference genome, but absent from the genome of the trace’s source, unassembled strain X. The insertion (black) interrupts the trace’s alignment (light gray) with the reference sequence. Overlapping traces were merged, identifying a unique indel. (Bottom) “Polymorphic insertion in strain X” represents an indel present in that strain (black) but absent from the reference genome. The sequence trace from strain X aligns well, but only partially, to the reference genome; another contiguous part of the trace does not align to the reference genome, identifying an indel variant (Supplemental Fig. 1). (B) An example of aligned genomic features in reference C57 and unassembled strains A and B genomic sequences, identifying various intermediate-length elements including polymorphic, nonpolymorphic, and reference sequences such as L1 retrotransposons. A sequence called an insertion in one strain might alternatively be considered a deletion from another.











