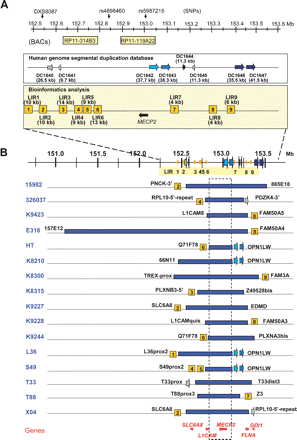
Bioinformatics analysis of the breakpoint regions and mapping of the duplications in the 16 patients. (A) Schematic overview of the repeat structures that coincide with duplication breakpoints in a 1-Mb region flanking MECP2 (152.55–153.55 Mb). Position and size of interchromosomal (DC1640 and DC1641) and intrachromosomal (DC1642, DC1643, DC1644, DC1645, DC1646, and DC1647) segmental duplications are depicted as reported in the Human genome segmental duplication database. In the Bioinformatics analysis box, LIR1 to LIR9 represent the breakpoint regions enriched for interspersed repeats with their lengths in brackets. The position of the BAC clones RP11-314B3 (152.63 Mb) and RP11-119A22 (152.90 Mb) used for FISH analysis is shown, as well as the position of DXS8087 (152.54 Mb) and the SNPs rs4898460 (152.83 Mb) and rs5987215 (153.05 Mb), used to investigate the origin of the duplications. (B) Duplication breakpoint mapping in the 16 patients (indicated on the left). Horizontal blue bars represent duplicated regions at Xq28. Size and location of each duplication were determined by iterative rounds of qPCR. Positions and sizes of all duplications were different. The colocalization of the breakpoint regions with the repeat structures, if present, is indicated with the name of the last duplicated qPCR primer set mentioned next to it (primer sequences can be found in Supplemental Table S1). The genes for which mutations are known to result in XLMR that also map in this region are shown at the bottom. The commonly duplicated region is boxed and includes only one XLMR gene, MECP2. Except for E316, all duplications lie within a 1.2-Mb region.











