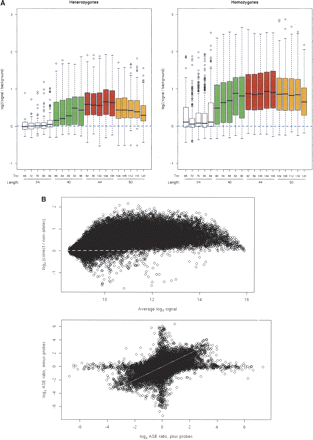
Optimization and validation of an array-based hybridization approach to allele-specific expression (ASE). (A) Optimization of feature design. (Left) For all cases in which a CEPH sample was heterozygous at a SNP, shown are the log2 ratios of the average of the two alleles at the SNP to the average of the two nonalleles, as a function of the length and Tm of the feature on the array. Probe lengths ranged from 29 bases to 55 bases (white = 29–39 nt, green = 35–45 nt, red = 40–49 nt, yellow = 45–55 nt), and relative melting temperatures of 68°C to 120°C (melting temperature increased from left to right within each color; see equation in design of array features in the Methods). (Right) For all cases in which a CEPH sample was homozygous at a SNP, shown are the log2 ratios of the allele present to the average of the two nonalleles, as a function of the length and Tm of the feature on the array. For both homozygotes and heterozygotes, the highest signal-to-noise was achieved by probe lengths of 40–49 nt. (B) Validation by allelotyping known alleles from cDNA of heterozygotes for a given SNP. (Top) The difference between the average log2 signal for the two alleles at the SNP and the average log2 signal for the two nonalleles, with results for all probes for a SNP combined. (Bottom) The estimated log2 ASE ratio at each SNP, as measured from the probes on the “plus” strand, against that measured from the probes on the “minus” strand.











