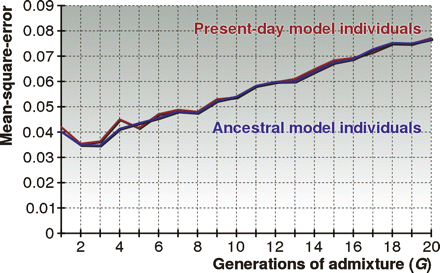
Performance of HAPAA when using ancestral versus present-day model individuals. For each G ∈ {1, 2, . . . , 20} we constructed (1) a set of unrelated ancestral individuals by randomly selecting 45 from each HapMap population and (2) 45 unrelated present-day model individuals for each population. Present-day individuals were generated over G generations of random mating using HapMap samples as templates. We tested on chromosome 22 on individuals admixed from the three populations over G generations using the mean-square-error metric.











