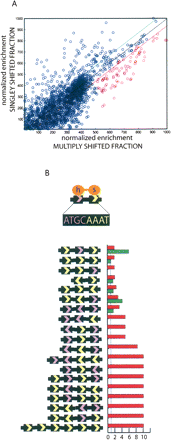
De novo motif identification. (A) For each oligo, singly bound (Y-axis) enrichment values were plotted against multiply bound enrichment (X-axis) values. POU5F1 contains two POU domains that recognize a bipartite signal as diagrammed in B. Half sites (ATGC, GCAT, AAAT, and ATTT) are counted in the entire set of oligonucleotidess, the set biased toward the singly bound state (above green line in A), and the set biased toward multiply bound (below the red line in A). Each permutation of half sites with more than twofold relative risk of being found in the multibound state versus the entire set is graphed. Red histogram bars mark relative risk (RR) of particular combination occurring in the multiply shifted fraction. Green bars for singly shifted fraction. Both measures are relative to the entire set and the blue dashed line marks zero enrichment (RR = 1).











