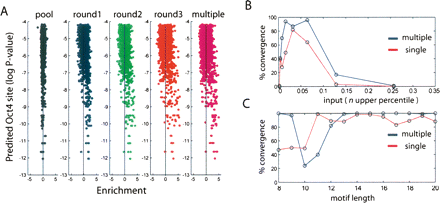
Comparison of octamer site prediction and POU5F1 binding. (A) POU5F1 sites were scored for each oligonucleotide as the log probability that a random sequence would fit the octamer binding model better than the highest scoring window (Y-axis) in the oligonucleotide and plotted against enrichment (X-axis). Vertical line represents mean enrichment for each experiment. (B) De novo motif identification was performed using Gibbs sampling trials with varying amounts of input that were ranked according to enrichment in round 1 (red) or the multiply bound fraction (blue). Successful trials that converged on motifs with the POU5F1 consensus (ATGCAAAT) were recorded on the Y-axis. (C) Using the top 3% of enriched oligonucleotides, the effect of motif length was examined.











