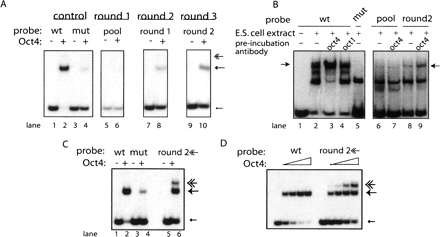
Enrichment for POU5F1 (Oct4) binding sites. (A) Perfect octamer, ATGCAAAT, containing oligonucleotide from the immunoglobulin heavy chain promoter (“wt” lanes 1,2) and its octamer scrambled control (“mut” lanes 3,4) were analyzed by EMSA with recombinant POU5F1(even number lanes) or no protein control (odd lanes). Mobility associated with singly bound oligonucleotides marked with an arrow; multiply bound oligonucleotides have been marked with a feathered arrow. Round 1 (lanes 5,6) of the POU5F1 enrichment was performed with the synthetic oligonucleotide pool as a probe. The singly shifted fraction was excised, reamplified, and used as a probe in lanes 7,8. The singly shifted fraction from round 2 was used as a probe in lanes 9,10. (B) EMSA performed using J1ES cell extract. Extract was preincubated with POU5F1 (Oct4) antibody (lanes 3,7,9) or with an antibody against a closely related POU2F1 (OCT1). Bands responsive to anti-POU5F1 antibodies are indicated with arrows. Lanes 6–9 display the oligonucleotide pool being shifted by ES extract. The starting oligonucleotide pool was used as probes for lanes 6,7, while lanes 8,9 use selected sequences from Round 2 (A, lane 8) of SELEX using the recombinant POU5F1. (C) Wild-type (wt), mutant, and the multiply shifted fraction of the oligonucleotide pool, excised (undetectable) from panel A, lane 8 (feathered arrow) were reamplified and used as a probe in an EMSA using recombinant POU5F1. (D) EMSA analysis performed using an increasing concentration gradient of recombinant POU5F1 protein on wild type and the multiply shifted fraction of the oligonucleotide pool.











