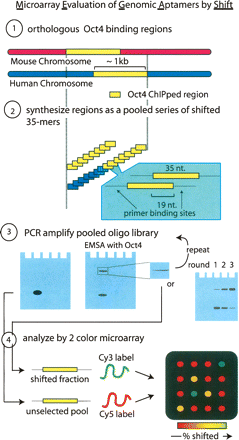
MEGAshift protocol. (Step 1) All orthologous genomic regions enriched in both human and mouse POU5F1 (Oct4) ChIP experiments were aligned and resynthesized (step 2) as a tiled contig of 35-mers flanked by universal primer binding sites. The human genomic region was extended to cover the union of this overlap. (Step 3) This pool was amplified with labeled primers migrates as a single band and was then used in an EMSA activity with recombinant POU5F1. The shifted band was excised, reamplified, and either reshifted or analyzed by cloning or microarray. (Step 4) Microarray analysis. Shifted and unselected fraction were reamplified and the T7 containing template used to generate Cy3 (shifted) or Cy5 (unselected) targets for the custom oligonucleotidearray.











