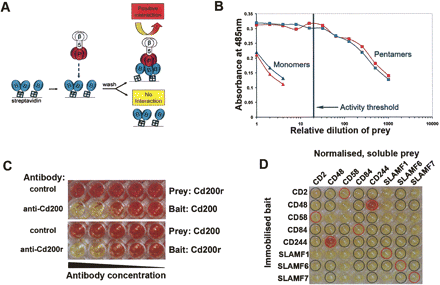
AVEXIS specifically detects low-affinity extracellular interactions with a low false-positive rate. (A) Schematic representation of AVEXIS. Biotinylated bait ectodomains (blue) are captured in an orientated manner on a streptavidin-coated microtiter plate and probed with soluble pentameric β-lactamase-tagged prey ectodomains (pink). Positive binding is detected by colorimetric enzymatic turnover of nitrocefin (yellow to red). (B) Pentamerization increases the avidity of prey ectodomains. Pentameric (squares) and monomeric (triangles) Cd200 (blue) and Cd200r (red) prey constructs were normalized for β-lactamase activity, serially diluted, and tested against the appropriate bait construct. The prey activity threshold used for AVEXIS is shown, corresponding to 50 units/mL. (C) AVEXIS interaction specificity. (Top panel) Low-affinity Cd200 (bait)–Cd200r (prey) interaction can be detected in the presence of a non-binding control antibody but is specifically blocked with an anti-Cd200 blocking monoclonal antibody; the blocking effect can be titrated by decreasing antibody concentration. The interaction specificity is independent of bait–prey orientation and can also be blocked with an anti-Cd200r antibody, bottom panel. (D) AVEXIS false-positive and -negative rate evaluation. An AVEXIS screening plate with bait proteins arrayed in rows and the prey proteins normalized to threshold levels in columns; positive binding results can be seen as red wells. Expected positive and negative wells are circled in red and black, respectively.











