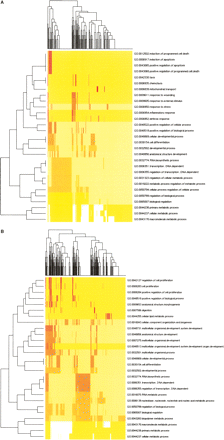
Clustering of over-represented Gene Ontology (GO) classes in predicted targets of differential microRNAs. Shown are heat map representations of GO (Ashburner et al. 2000) terms over-represented among predicted cooperative targets (Y-axis) of hESC-enriched miRNAs (A) and EB-enriched miRNAs (B). All genes with statistically over-represented GO annotations were included (P < 0.01, X-axis) as identified by GoStat (Beissbarth and Speed 2004). GO terms common to both sets of genes were those involved in transcriptional regulation, differentiation, and development. Those GO terms unique to hESC-enriched miRNA targets were associated with programmed cell death, response to stress, and cell motility while those unique to EB-enriched miRNA targets describe various aspects of cell proliferation regulation as well as nucleotide and nucleic acid metabolism.











