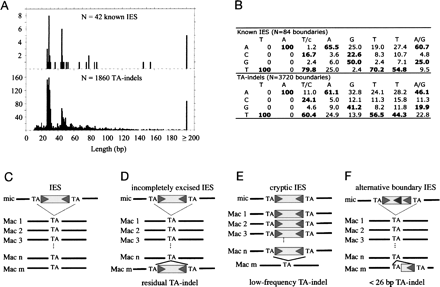
Sequence analysis of TA-indels and comparison with known IESs. (A) Length distribution of TA-indels and known IESs in P. tetraurelia. (B) Consensus sequence and frequency matrix of the 8-bp motif at the boundaries of TA-indels and of known IESs. Nucleotide frequencies that are higher than the genome average are indicated in bold. (C–F) Putative relationships between IESs and TA-indels. The IES is drawn as a box flanked by two TA repeats; triangles indicate the additional six nucleotides that define the consensus for the terminal inverted repeats. IESs that are excised with 100% efficiency are totally absent from the macronucleus, and therefore cannot be detected as TA-indels in our analyses (C). TA-indels may correspond to IESs for which some copies remain unexcised (D) or to cryptic IESs that are excised at low frequency (E). Some TA-indels (and notably those <26 bp) may correspond to IESs with alternative boundaries (F). Some TA-indels may also correspond to imprecise deletion events (data not shown).











