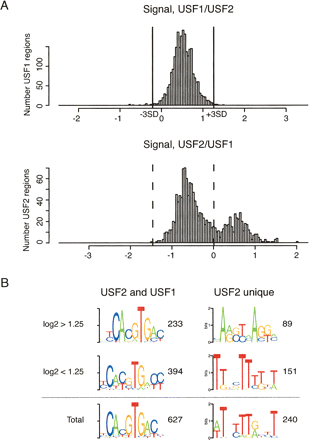
USF2 binds a subset of unique targets. (A) For USF1 (top) and USF2 (bottom) the log2 ratios between USF1/USF2 and USF2/USF1 signals were calculated (X-axes) and shown in histograms. The vertical gray lines demarcate the regions deviating >±3 SD from the median of the USF1/USF2 distribution. (B) USF2-bound regions were divided based on the log2 USF2/USF1 signal ratios in A, in those with ratios higher than 0.5 (USF2 unique) or lower than –0.5 (USF2–USF1 heterodimers). Enriched motifs were identified ab initio for all USF2-unique and USF2–USF1 regions, and after dividing them in groups based on log2 enrichment signals.











