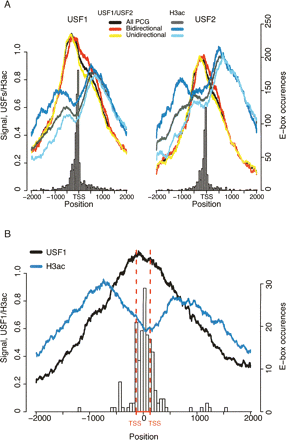
Architecture of USFs bound promoters. (A) PCGs bound by USF1 (left) or USF2 (right) within ±1 kb of their TSS. The ChIP-chip signals (in log2 scale, right Y-axes) of USF1 or USF2 and H3ac were plotted around TSS. Profiles are presented for all USF1- or USF2-bound PCG (all), bidirectional PCGs (at least one anti-sense PCG or human mRNA TSS within 1 kb of their TSS), or unidirectional PCGs (no anti-sense PCG, human mRNA, or spliced EST TSS within 1 kb of their TSS). Histograms of perfect matches to E-box consensus sequence (CACGTGAC, left Y-axes) are presented for all USF1- or USF2-bound genes in 50-bp windows. (B) All PCGs in a non-overlapping bidirectional conformation bound by USF1 within ±1 kb of their TSSs were selected. The binding profiles (left Y-axes) of USF1 and H3ac were plotted around 2 kb of the middle position between the two transcripts in each bidirectional pair. The red dotted lines show the median size of the intergenic region between TSSs in each bidirectional pair. The histogram shows E-box consensus occurrences (right Y-axes).











