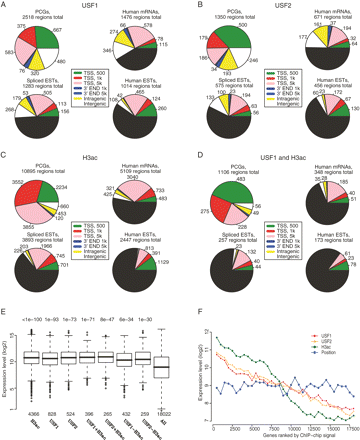
Mapping of USFs and H3ac binding sites to different gene and transcript annotations and correlation with expression levels. Pie charts are presented for USF1 (A), USF2 (B), H3ac (C), and USF1+H3ac (regions enriched both for USF1 and H3ac) (D). In each case, mapping of sites to the different genomic location was first established for known genes/Ensembl/RefSeq; subsequently for RNA genes, human mRNAs/spliced ESTs; and lastly for human ESTs. In each annotation, the regions mapping within 500 bp or 1 kb of TSS were not mapped in the following annotation and are indicated by the black portions in the charts. The number of regions corresponding to each genomic location category is presented next to the pie chart fractions. (E) The expression level of ∼18,000 genes was collected from Gene Expression Omnibus (Supplemental methods). The box plots show the overall expression (in log2 scale) for all 18,000 genes and for the genes bound by the proteins indicated at the bottom of the graph within ±1 kb of their TSS. P-values from two-tailed t-tests between each group of genes compared with all genes are given above the box plots. (F) Genes were ranked in descending order according to the maximal ChIP-chip signals within ±1 kb from TSS for USF1, USF2, or H3ac experiments. As a control, we also ranked genes according to their genomic positions. Within each ranking (X-axes), we plotted the mean expression levels (Y-axes) of groups of 500 genes.











