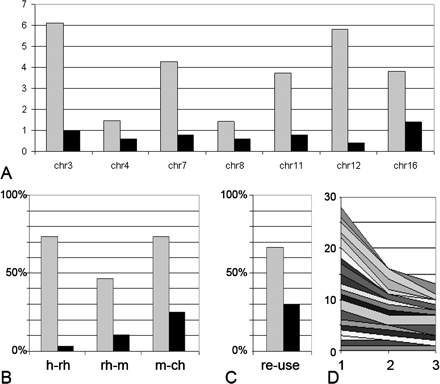
Frequencies of breaks within regions surrounding the TBSDs during mammalian evolution. (A) Total number of human–rhesus, rhesus–mouse, and mouse–chicken synteny breaks per megabase within regions surrounding the TBSDs (gray) and within random 500-kb sites (black) taken from each 10 Mb of the particular chromosome (chr 3, chr 4, etc) sequence. (B) Percentage of regions, which had human–rhesus (h-rh), rhesus–mouse (rh-m), and mouse–chicken (m-ch) synteny breaks, counted from total number of analyzed 500-kb regions, which surrounded either TBSDs (gray) or random sites taken at each 10 Mb of TBSD containing chromosome sequences (black). (C) Frequency of reuse for synteny breakpoint region at TBSD (gray) and at random synteny break (black). Synteny breakpoint region is defined as a 500-kb region containing a minimum of one break in either human–rhesus or in rhesus–mouse or in mouse–chicken branch. Reuse of the synteny breakpoint region is defined as the occurrence of breaks in at least two of three mentioned branches during evolution. (D) Gradual decrease in frequency of rhesus–mouse and mouse–chicken synteny breaks with increase of distance from TBSD sites. Cumulative area shows the number of breaks within the 1st, 2nd, and 3rd sequence windows (point 1, 2, and 3, respectively) from different TBSD locations. For each TBSD location, windows 1, 2, and 3 represent successive sequence windows equal in size to TBSD, which flank TBSD (1), located next to both flanks (2), and most distant windows (3).











