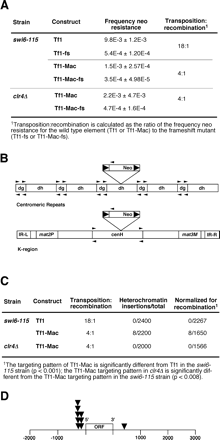
Target site choice of the Tf1 retrotransposon in S. pombe is altered by adding the MAGGY CHD to integrase. (A) Transposition and cDNA recombination frequencies of Tf1 and Tf1-Mac. The MAGGY CHD was fused to the C terminus of Tf1 integrase, creating Tf1-Mac. Tf1-Mac transposes, but at a lower frequency than wild-type Tf1. Integrase frameshift mutants (Tf1-fs and Tf1-Mac-fs) abolish Tf1 or Tf1-Mac integration, and therefore serve to infer frequencies of cDNA recombination. (B) A screen for insertions of Tf1 and Tf1-Mac in heterochromatin. A diagram of S. pombe centromeric repeats and mating type region depicting the location of the PCR primers used to identify heterochromatic Tf1 insertions. (C) Targeting of Tf1-Mac to heterochromatin. Centromeric heterochromatin in S. pombe, which is enriched in histone H3 methyl-K9, becomes a target site for Tf1-Mac in the swi6-115 strain. In the swi6-115 strain, there are eight integration events into heterochromatin out of 2200 Tf1-Mac transposition events. This number of transposition events includes elements incorporated into the genome by either integration or recombination. The number that arose specifically by integration was inferred by normalizing the data using the value for the ratio of transposition to recombination (4:1). The normalized heterochromatin targeting frequency is eight out of 1650 integration events. Tf1-Mac does not target into heterochromatin in the clr4Δ strain, which lacks H3 methyl-K9. (D) Native Tf1 target specificity is retained by the Tf1-Mac construct. Fusion of the MAGGY CHD to Tf1 does not abolish Tf1’s native targeting specificity in the swi6-115 strain. Sites of insertion for nine randomly selected Tf1 integration events were determined by inverse PCR. Eight were found within promoters and one near a transcription terminator, consistent with previously documented Tf1 targeting patterns (Behrens et al. 2000; Singleton and Levin 2002).











