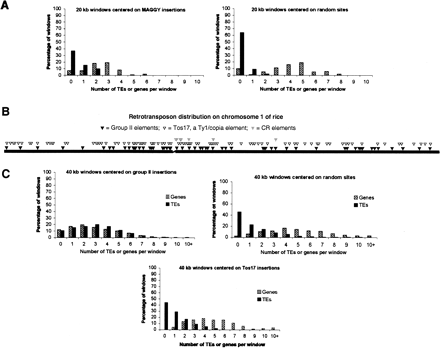
Retrotransposons with CHDs and CR motifs are preferentially located in gene-poor, transposon-rich regions of their host genomes. (A) The genomic distribution of M. grisea MAGGY (group I) insertions. For each of the 75 MAGGY insertions in the completed genome sequence (Dean et al. 2005), 20-kb windows at the site of insertion (10 kb upstream and downstream) were surveyed for genes and transposable elements. Seventy-five randomly selected sites were similarly surveyed as a control. (B) The distribution of group II, CR, and Tos17 insertions on chromosome 1 of O. sativa. Insertions of group II chromoviruses and Tos17 are distributed along the chromosome arms; however, as indicated in C below, these two element families occupy different genomic contexts. Only a few CR elements are found in the assembled rice genome sequence and these are near the pericentromeric regions (gap in chromosome). Many other CR insertions have been revealed by FISH analysis or are present in the recent sequence of the centromere from chromosome 8 (Cheng et al. 2002; Nagaki et al. 2004). (C) Os (class II) insertions are in transposon-rich, gene-poor regions scattered across the 12 rice chromosomes. For each Os insertion site, a 40-kb window (20 kb upstream and 20 kb downstream) was surveyed for genes and transposable elements. Windows containing Os insertions have more transposable elements and fewer genes than random windows or windows centered on Tos17 insertions, a retrotransposon that integrates preferentially into gene-rich regions of the rice genome (Miyao et al. 2003).











