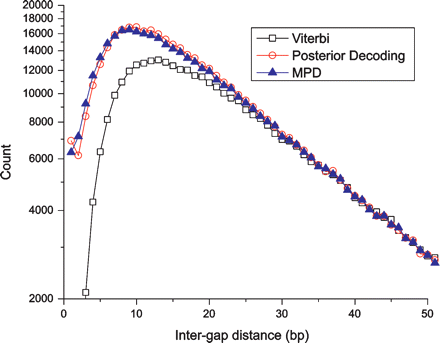
Posterior decoding shows fewer alignment biases. Shown are the intergap distance histograms for alignments obtained by Viterbi decoding (open squares), posterior decoding (open circles) and MPD (filled triangles), applied on the Full model. The scarcity of closely spaced gaps, resulting from gap attraction, is apparent for all decoding algorithms, but is much less pronounced for posterior decoding and MPD than for Viterbi decoding.











