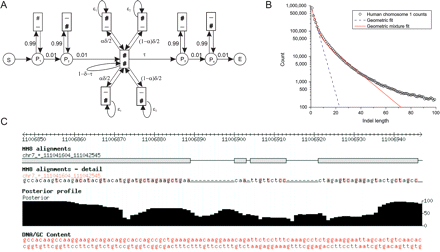
Topology of the pair HMM for probabilistic alignments. (A) The model is implemented as a pair HMM with a match state (center) surrounded by delete (top) and insert (bottom) states. Hash signs (#) signify emissions, dashes (–) represent no emission (rather than the emission of a gap character); circles represent silent states and are included for clarity, and arrows represent allowed transitions. Paths through this HMM correspond to alignments (and dash signs then represent gap characters). Local alignments were computed by surrounding the core HMM by two pairs of “padding” states (P1 to P4) allowing the alignable portion of the sequences to be embedded in nonhomologous sequence. Note that the model allows a single pass through the central pair HMM, and padding sequence is allowed at both ends of the alignment only. (B) The observed indel-length spectrum in BLASTZ human–mouse alignments (right, circles) is better approximated by a mixture of two geometric distributions (red solid line) than by a single geometric distribution (corresponding to affine-gap scores; blue dashed line). This mixture distribution is implemented by duplicating the insert and delete states. Parameters of the model are: δ, the indel probability per aligned site; ε1 and ε2, the parameters governing the indel length distribution; α, the geometric mixture coefficient, τ, the alignment length parameter. (C) Screenshot of the alignment browser, showing a marginalized posterior decoding (MPD) alignment computed using this model, together with posterior column probabilities. Alignments generally contain columns with low posterior probability, indicating regions where competing alignments contribute a significant fraction of the total likelihood.











