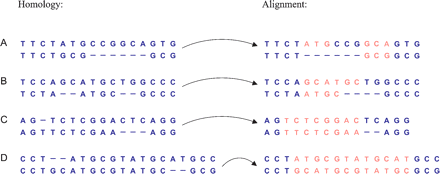
Three types of alignment bias. Alignment algorithms are consistently biased toward likely distributions of indels across sequences, despite the occurrence of less-likely configurations at low frequencies. The figure shows four pairs of sequences with their homologies (left) and corresponding most-likely alignments (right), with wrongly aligned bases highlighted. We distinguish between three types of bias: gap wander (A), caused by spurious high-sequence similarity at nonhomologous sites; gap attraction (B,C), occurring when two indels have little separation, and gap annihilation (D), which occurs when two indels of equal size but opposite signature are found near to each other, favoring explanations without indel events.











